Page Not Found
Page not found. Your pixels are in another canvas.
A list of all the posts and pages found on the site. For you robots out there is an XML version available for digesting as well.
Page not found. Your pixels are in another canvas.
About Martin Skarzynski
This is a page not in th emain menu
Updated:
This post will show up by default. To disable scheduling of future posts, edit config.yml and set future: false.
A web application that can predict the next word based on text input from the user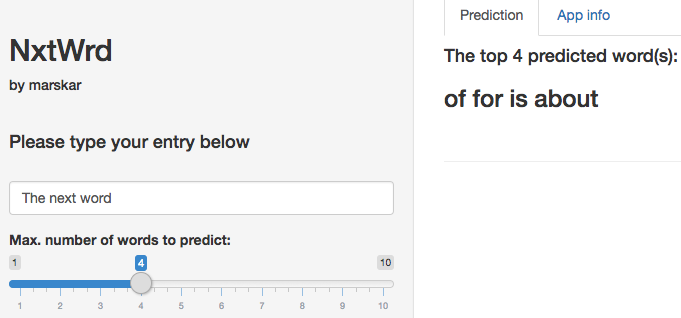
I am currently working with Professor Elizabeth Platz to study lung cancer risk using genomic and epidemiologic data from ARIC.
To view a slideshow about this project, please click here.
I am currently working with Professor Kasper Daniel Hansen to develop analysis and probe design tools for the LISH assay.
Published in Cancer Research, 2014
Targeting the IgM receptor on Chronic Lymphocytic Leukemia with an Antibody Drug Conjugate
Download here
Published in Bioconjugate Chemistry, 2015
Targeting CD22 and mesothelin on cancer cells with recombinant immunotoxins
Download here
Published in Frontiers in Public Health, 2015
Discussion of HPV vaccination health disparities in the US
Download here
Published in Clinical Cancer Research, 2016
Targeting Chronic Lymphocytic Leukemia with an Anti-CD20 Antibody and a Brutons Tyrosine Kinase (BTK) Inhibitor
Download here
Updated:
This is a description of your talk, which is a markdown files that can be all markdown-ified like any other post. Yay markdown!
Updated:
This is a description of your conference proceedings talk, note the different field in type. You can put anything in this field.
Graduate Course, Foundation for the Advanced Education in the Sciences, 2018
Between Electronic Medical Records and Electronic Health Records, PubMed, and collections of biomedical grant applications, there exist large quantities of medical information stored in databases waiting to be explored. Besides tables of numbers, medical records also contain a great amount of free-text paragraphs that are comprehensible to human readers but challenging to computers. Text mining is an interdisciplinary area that primarily combines advances in Natural Language Processing (NLP), Information Retrieval (IR), and Machine Learning (ML) to help the computers understand human written language and thus extract medical and clinical information from free-text records. This class aims to introduce fundamental subjects in text mining such as tokenization, named entity recognition (NER), grammars, parsing, relation extraction, and document classification. The class is oriented towards hands-on experience with Python and Natural Language Toolkit (NLTK). Thank you to DataCamp for offering free full access to all of their awesome content to all of our students! Thank you to JetBrains for offering free full access to their All Product Pack, including PyCharm Professional, to all of our students!
Graduate Course, Foundation for the Advanced Education in the Sciences, 2018
In this course, participants learn basic pharmacology theory with introductory statistics using a popular open source software program (R Studio) that is capable of conducting pharmacokinetic (PK) exposure and pharmacodynamic (PD) response analyses from example clinical trial data. Ultimately, the framework of analyzing exposure/response relationships will be demonstrated in order to make decisions to guide drug development. For access to the course materials (code and slides), please visit the April 2017 BioTech 84 repository on GitHub.
Online Course, DataCamp, 2019
The decisions we make in life are guided by our principles. No one is born with a life philosophy, instead everyone creates their own over time. In the Creating Robust Python Workflows course, you will develop a set of principles for your data science and software development projects. These principles will save time, prevent frustration, and build your confidence as a data scientist and software developer. In addition to best practices in the Python programming language, You will learn to leverage hidden gems in the Python standard library and well-known tools from Python’s excellent ecosystem, such as pandas and scikit-learn. The time you invest in Creating Robust Python Workflows will yield dividends for you and others throughout your career. Your colleagues, community members, and future self will thank you.
Graduate Course, Foundation for the Advanced Education in the Sciences, 2019
The Introduction to Python (BIOF309) course is designed for non-programmers, biologists, or those without specific knowledge of Python to learn how to write Python programs that expand the breadth and depth of their research. Python is a free, open-source and powerful programming language that is easy to learn. Students will learn to use Jupyter Notebooks and the PyCharm integrated development environment (IDE). Thank you to JetBrains for offering free full access to their All Product Pack, including PyCharm Professional, to all of my students! Week by week we will slowly build up our skills and understanding of programming and the Python language. There will be in-class demonstrations, using PyCharm and Jupyter Notebooks, and activities to be completed outside of class, mostly using DataCamp, for you to practice and learn at your own pace. Thank you to DataCamp for offering free full access to all of their awesome content to all of my students! For a quick idea of the course, please take a look at the Fall 2018 Syllabus. For access to the course materials and more information about the course, please visit the Fall 2018, Spring 2018, and Fall 2017 BIOF309 repositories on GitHub.
Graduate Course, Foundation for the Advanced Education in the Sciences, 2019
Machine learning is a computational field that consists of techniques allowing computers to learn from data and make data-driven predictions or decisions. The ability to implement machine learning approaches appropriately and intelligently is a crucial component of data analysis. The Applied Machine Learning (BIOF509) course provides a broad practical introduction to machine learning concepts, analysis design, and implementation.The course will give a broad and conceptual overview of the most popular machine learning algorithms, followed by examples of how and when to apply them to real data. Best practices in designing machine learning analyses will be emphasized and reviewed, along with how to avoid common pitfalls and how to interpret analysis results. Through homework and in-class assignments, students will implement machine -earning models in Python, utilizing state-of-the-art machine learning Python packages, such as scikit-learn and tensorflow. Algorithms that will be covered include, but are not limited to linear and logistic regression, random forest, K-means clustering, and deep learning. Note that the course emphasizes hands-on application of algorithms, and mathematical derivation will not be reviewed. Further, depending on the students’ familiarity with Python, completing the weekly homework assignments can take one to four hours. The course will culminate in a short research project utilizing machine learning to analyze either the student’s own dataset or a public dataset that the student chooses.
Enterprise Training Course, General Assembly, 2019
The Tech Excellence Data Science (TEDS) course is part of the Data Science 5000 (DS5K) initiative. DS5K has the ambitious goal of training 5000 Data Scientists at Booz Allen Hamilton in the 5 years. The course is open to any Booz Allen Hamilton employee and is developed in partnership between Booz Allen Hamilton and General Assembly, a pioneer in tech education and career transformation. For more information on the course, please take a look at the GitHub repo for Tech Excellence Data Science (TEDS).